Position/Role: GSAT M.Sc. Student
Research Interests: Molecular Epigenetics, Bioinformatics, DNA Methylation
Contact: tony.hui[at]ubc.ca
Major research activities/interests:
DNA methylation is an epigenetic mark that controls gene expression in mammals. Current methods used to detect DNA methylation suffer from a lack of sensitivity: these methods require a lot of genomic DNA (often too much DNA) as input. Assessing DNA methylation patterns in rare cell populations or assessing DNA methylation heterogeneity within a cell population are currently impossible.
My undergraduate honours thesis involved developing a highly sensitive method for DNA methylation detection that requires only a single-cell as input. Currently, we have a protocol finalized, and are working through the challenges in data analysis from a bioinformatics perspective.
In collaboration with research labs all across UBC, our lab plans on using this newly developed method to study hematopoietic stem cells and individual cancer cells to better understand their DNA methylation patterns and how they contribute to development/disease.
Publications:
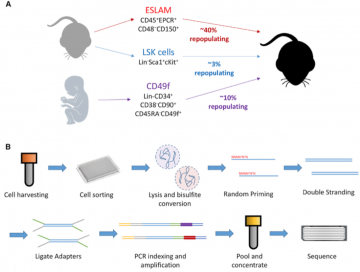
This Stem Cell Reports paper present an analytical strategy (PDclust) developed in Dr. Hirst lab to define single-cell DNA methylation states through pairwise comparisons of single-CpG methylation measurements:
Hui et al., High-Resolution Single-Cell DNA Methylation Measurements Reveal Epigenetically Distinct Hematopoietic Stem Cell Subpopulations, Stem Cell Reports (2018), https://doi.org/10.1016/j.stemcr.2018.07.003
Knox, Rachel, Joanne E. Nettleship, Veronica T. Chang, Zhao Kun Hui, Ana Mafalda Santos, Nahid Rahman, Ling-Pei Ho, Raymond J. Owens, and Simon J. Davis. “A streamlined implementation of the glutamine synthetase-based protein expression system.” BMC biotechnology 13, no. 1 (2013): 74.
